I failed to install DBD::mysql. This time I had the dev files and everything.
FAIL Installing DBD::mysql failed. See /home/pmg/.cpanm/build.log for details.
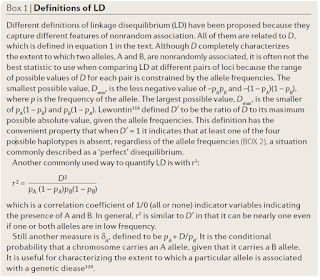
$ cpanm DBD::mysql
[...]
DBD::mysql::db do failed: alter routine command denied to user ''@'localhost' for routine 'test.testproc' at t/80procs.t line 41.
Looking at this line of code:
my $drop_proc= "DROP PROCEDURE IF EXISTS testproc";
ok $dbh->do($drop_proc);
And looking at my
mysql db table I can see that there is no privileges for alter or execute procedures
mysql> select * from db\G
Host: localhost
Db: test
User:
Select_priv: Y
Insert_priv: Y
Update_priv: Y
Delete_priv: Y
Create_priv: Y
Drop_priv: Y
Grant_priv: N
References_priv: Y
Index_priv: Y
Alter_priv: Y
Create_tmp_table_priv: Y
Lock_tables_priv: Y
Create_view_priv: Y
Show_view_priv: Y
Create_routine_priv: Y
Alter_routine_priv: N # <<====
Execute_priv: N # <<====
Event_priv: Y
Trigger_priv: Y
mysql> show grants for ''@localhost;
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
| Grants for @localhost |
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
| GRANT USAGE ON *.* TO ''@'localhost' |
| GRANT SELECT, INSERT, UPDATE, DELETE, CREATE, DROP, REFERENCES, INDEX, ALTER, CREATE TEMPORARY TABLES, LOCK TABLES, CREATE VIEW, SHOW VIEW, CREATE ROUTINE, EVENT, TRIGGER ON `test`.* TO ''@'localhost' |
| GRANT SELECT, INSERT, UPDATE, DELETE, CREATE, DROP, REFERENCES, INDEX, ALTER, CREATE TEMPORARY TABLES, LOCK TABLES, CREATE VIEW, SHOW VIEW, CREATE ROUTINE, EVENT, TRIGGER ON `test\_%`.* TO ''@'localhost' |
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
4 rows in set (0.00 sec)
Add permision to anyone from localhost to test
mysql> grant ALL on test.* to ''@'localhost';
mysql> show grants for ''@localhost;
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
| Grants for @localhost |
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
| GRANT USAGE ON *.* TO ''@'localhost' |
| GRANT ALL PRIVILEGES ON `test`.* TO ''@'localhost' |
| GRANT SELECT, INSERT, UPDATE, DELETE, CREATE, DROP, REFERENCES, INDEX, ALTER, CREATE TEMPORARY TABLES, LOCK TABLES, CREATE VIEW, SHOW VIEW, CREATE ROUTINE, EVENT, TRIGGER ON `test\_%`.* TO ''@'localhost' |
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------+
4 rows in set (0.00 sec)
mysql> select * from db\G
*************************** 1. row ***************************
Host: localhost
Db: test
User:
Select_priv: Y
Insert_priv: Y
Update_priv: Y
Delete_priv: Y
Create_priv: Y
Drop_priv: Y
Grant_priv: N
References_priv: Y
Index_priv: Y
Alter_priv: Y
Create_tmp_table_priv: Y
Lock_tables_priv: Y
Create_view_priv: Y
Show_view_priv: Y
Create_routine_priv: Y
Alter_routine_priv: Y ## <<<<==== OK
Execute_priv: Y ## <<<<==== OK
Event_priv: Y
Trigger_priv: Y